Abstract
Background. Insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2) is implicated in various cancers, but its role in modulating ferroptosis and tumor cell behavior in non-small cell lung cancer (NSCLC) remains unclear.
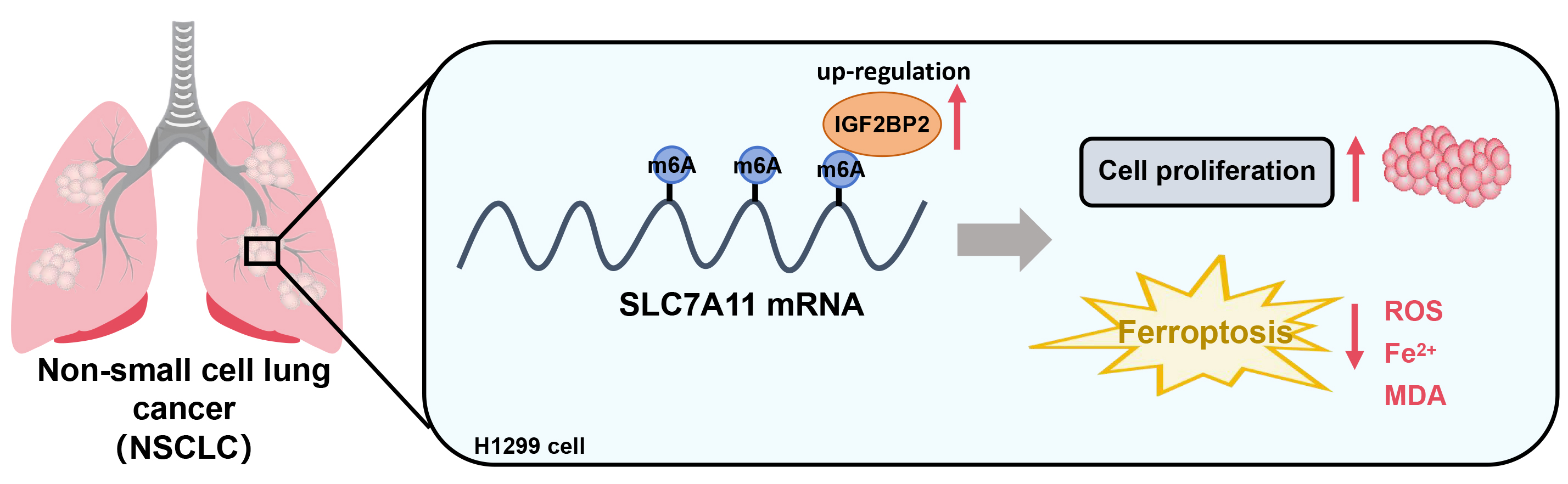
Objectives. This study aimed to investigate how IGF2BP2-mediated N6-methyladenosine (m6A) modification of solute carrier family 7 member 11 (SLC7A11) affects ferroptosis and NSCLC cell viability.
Materials and methods. NSCLC H1299 cells were transfected with either IGF2BP2 or SLC7A11 plasmids and corresponding siRNAs. Expression levels of IGF2BP2, SLC7A11 and ferroptosis markers were analyzed using reverse transcription real-time quantitative polymerase chain reaction (RT-qPCR) and western blot. Cell viability was assessed using the Cell Counting Kit-8 (CCK-8) assay. Reactive oxygen species (ROS) and lipid peroxidation levels were measured with flow cytometry and biochemical kits. The RNA immunoprecipitation (RIP) and mRNA stability assays were utilized to explore the interaction between IGF2BP2 and SLC7A11.
Results. IGF2BP2 expression was significantly upregulated in H1299 cells. Overexpression of IGF2BP2 enhanced cell viability and decreased ferroptosis, whereas its knockdown resulted in reduced cell viability and increased ferroptotic activity. IGF2BP2 enhanced SLC7A11 mRNA stability through m6A modification, and SLC7A11 overexpression reversed the effects of IGF2BP2 knockdown. This interaction increased cell viability and reduced ROS and lipid peroxidation.
Conclusions. IGF2BP2 plays a critical role in NSCLC by stabilizing SLC7A11 mRNA via m6A modification, promoting cell proliferation and suppressing ferroptosis. Targeting the IGF2BP2–SLC7A11 axis may offer a promising therapeutic strategy for NSCLC.
Key words: non-small cell lung cancer, SLC7A11, ferroptosis, IGF2BP2, m6A modification
Background
Non-small cell lung cancer (NSCLC), the most prevalent form of lung cancer, constitutes nearly 85% of lung cancer diagnoses. Besides, NSCLC is characterized by a complex interplay of genetic and metabolic factors that contribute to lung cancer progression and resistance to treatment.1 Insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2) is pivotal in the context of NSCLC, serving as a regulatory protein related to cancer biology due to its effects on cellular proliferation, metabolism and survival.2
IGF2BP2 exerts its biological functions primarily by binding to the mRNAs of various genes, thereby regulating their stability and translation.3 As a critical target of IGF2BP2, solute carrier family 7 member 11 (SLC7A11) is a crucial transporter in the cellular antioxidant system that maintains intracellular glutathione levels.4, 5 Ferroptosis, a regulated cell death process induced by oxidative stress and characterized by lipid peroxidation, has attracted growing attention in cancer research due to its potential as a therapeutic target.6 SLC7A11 acts greatly in preventing ferroptosis via promoting the uptake of cystine, which is essential for glutathione synthesis. Glutathione serves as a crucial antioxidant, safeguarding cells against lipid peroxidation and oxidative damage. The capability of cancer cells to evade ferroptosis through metabolic alterations, including the upregulation of glutaminolysis, is considered a hallmark of cancer cell survival and proliferation.7 The conversion of glutamine into α-ketoglutarate was conducted through this metabolic pathway, thereby fueling the tricarboxylic acid cycle. Additionally, such a pathway is often enhanced in cancer to meet the increased demands of rapidly dividing cells and contribute to the perturbation of redox balance.8, 9 Given the importance of SLC7A11 in maintaining redox homeostasis and preventing ferroptosis, it was specifically chosen for this study.
Objectives
In this context, the present study was designed to elucidate the roles of IGF2BP2 and SLC7A11 in regulating the fate of NSCLC cells, with a particular focus on their contributions to cell viability and resistance to ferroptosis.
We hypothesized that IGF2BP2 was upregulated in NSCLC cells and this upregulation confers a growth advantage through the stabilization of SLC7A11 mRNA, enhanced antioxidant defense and metabolic shifts favoring cell survival.10, 11, 12 The purpose of this study was to explain the regulatory functions of IGF2BP2, examining its impact on the post-transcriptional control of critical genes and its broader effects on the NSCLC cell’s metabolic state.
Materials and methods
Cell culture and transfection
Normal human bronchial epithelial (HBE) cells (CRL-2741) and NSCLC H1299 cells (CRL-5803) were provided by the American Type Culture Collection (ATCC; Manassas, USA). The culture of HBE cells and H1299 cells was conducted in Dulbecco’s modified Eagle’s medium (DMEM)/Nutrient Mixture F-12 medium (cat. No. #11320033; Gibco, Waltham, USA) and Roswell Park Memorial Institute (RPMI) 1640 medium (cat. No. #11875093; Gibco), respectively. Both media contained 1% penicillin/streptomycin (cat. No. #15140122; Gibco) as well as 10% fetal bovine serum (FBS; cat. No. #10082147; Gibco). Cells were incubated at 37°C in a humidified atmosphere containing 5% CO₂.
For transfection, H1299 cells were seeded into 6-well plates to reach 70–80% confluency. Transfections were performed using Lipofectamine 2000 (cat. No. #11668019; Invitrogen, Waltham, USA) according to the manufacturer’s instructions. The following experimental groups were established:
SLC7A11 group: transfected with SLC7A11 pcDNA3.1 plasmid; IGF2BP2 group: transfected with IGF2BP2 pcDNA3.1 plasmid; Vector group: transfected with empty vector (control for plasmid transfection); si-IGF2BP2 group: transfected with siRNA targeting IGF2BP2
(sense: 5’-GGGACCAAGAUAACAAUCUTT-3’,
anti-sense: 5’-AGAUUGUUAUCUUGGUCCCTT-3’);
si-NC group: transfected with negative control siRNA
(5’-CAACAAGAUGAAGAGCACCAA-3’,
anti-sense: 5’-UUGGUGCUCUUCAUCUUGUUG-3’;
100 nM final concentration).
For each group, the corresponding controls (empty vector or negative control siRNA) were included to ensure the specificity of the observed effects. Cells were harvested 48 h post-transfection for further analysis. Sangon Biotech Co., Ltd. (Shanghai, China) was responsible for designing and synthesizing the aforementioned plasmids, and the siRNA fragments were provided by GenePharma (Shanghai, China).
RT-qPCR
TRIzol reagent (cat. No. #15596018; Invitrogen) was utilized for extracting total RNA from cells according to the manufacturer’s protocol. A NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, USA) was adopted to assess the quantity and quality of RNA. Besides, as described by the manufacturer’s guidelines, the synthesis of cDNA was conducted using total RNA (1 µg) through the High-Capacity cDNA Reverse Transcription Kit (cat. No. #4368814; Applied Biosystems, Waltham, USA). Reverse transcription real-time quantitative polymerase chain reaction (RT-qPCR) was performed using SYBR Green PCR Master Mix (cat. No. #4309155; Applied Biosystems) on a QuantStudio 5 Real-Time PCR System (Applied Biosystems). Primer sequences applied were as follows:
IGF2BP2:
Reverse 5’-TGGTAGGTGGTCTCGGTGTT-3’,
Forward 5’-AGAAGATGTGGAGGAGGCTT-3’;
GAPDH (internal control):
Reverse 5’-GAAGATGGTGATGGGATTTC-3’,
Forward 5’-GAAGGTGAAGGTCGGAGTC-3’;
SLC7A11:
Reverse 5’-AGACTCCCCTCAGTAAAGTGAC-3’,
Forward 5’-TCTCCAAAGGAGGTTACCTGC-3’.
Relative gene expression was analyzed using the 2−ΔΔCt method, with GAPDH served as the internal control to normalize the results.
Western blot
Cell were lysed with ice-cold radioimmunoprecipitation assay (RIPA) buffer (cat. No. #R0278; Sigma-Aldrich, St. Louis, USA) supplemented with phosphatase inhibitors (cat. No. #P0044; Sigma-Aldrich) as well as protease (cat. No. #P8340; Sigma-Aldrich). Protein concentrations were measured using the Bicinchoninic Acid (BCA) Protein Assay Kit (cat. No. #23225; Pierce, Rockford, USA). Proteins (30 μg) were separated on 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene fluoride (PVDF) membranes (cat. No. #IPVH00010; MilliporeSigma, St. Louis, USA). Membranes were blocked with 5% skimmed milk for 1 h at room temperature and incubated overnight at 4°C with the following primary antibodies: IGF2BP2 (cat. No. #14672S, 1:1,000; Cell Signaling Technology (CST), Danvers, USA), SLC7A11 (cat. No. #ab175186, 1:1,000; Abcam, Cambridge, UK), GPX4 (cat. No. #ab125066, 1:1,000; Abcam), β-actin (cat. No. #A1978, 1:5,000; Sigma-Aldrich), and Acyl-CoA synthetase long-chain family member 4 (ACSL4) (cat. No. #ab155282, 1:1,000; Abcam). Upon rinsing, the membranes were subjected to 1-h incubation with horseradish peroxidase (HRP)-conjugated secondary antibodies (cat. No. #NA931, 1:5,000 for anti-rabbit; cat. No. #NA934, 1:5,000 for anti-mouse; GE Healthcare, Chicago, USA) at ambient temperature. Visualization of the protein bands was achieved using the SuperSignal West Pico PLUS Chemiluminescent Substrate (cat. No. #34580; Thermo Fisher Scientific), followed by imaging on a ChemiDoc XRS+ system (Bio-Rad, Hercules, USA).
Cell viability assay
Cells (5 × 103 cells/well) were seeded into 96-well plates and allowed to adhere overnight. Following treatments, 100 µL of fresh medium containing MTT reagent (0.5 mg/mL, cat. No. #M2128; Sigma-Aldrich) was added and incubated for 4 h. The formazan crystals were dissolved in 100 µL of dimethyl sulfoxide. A microplate reader (Bio-Tek; Winooski, USA) was utilized for measuring the absorbance at 570 nm, with a reference wavelength of 690 nm. Untreated cells served as a control for baseline viability.
ROS detection
The DCFDA Cellular ROS Detection Assay Kit (cat. No. #ab113851; Abcam) was employed for detecting the intracellular ROS levels. Cells (1 × 104 cells/well) were planted in a 96-well plate for cultivation until reaching 70–80% confluency. Upon rinsing with phosphate-buffered saline (PBS), cells was incubated with 25 µM DCFDA in a serum-free medium for 45 min at 37°C in the dark. Fluorescence intensity was measured using a microplate reader (Bio-Tek; excitation: 485 nm, emission: 535 nm). Negative controls (untreated cells) were used to set the baseline fluorescence levels.
Measurement of malondialdehyde and Fe2+ levels
Following treatments, the collected cells were rinsed with pre-cooled PBS. Upon centrifugation (3,000 rpm, 4°C, 15 min), the supernatant was collected to assess the levels of Fe2+ and malondialdehyde (MDA) using the Iron Assay Kit (cat. No. 3100865; Sigma-Aldrich) and MDA Assay Kit (cat. No. MAK085; Sigma-Aldrich), respectively, as per the manufacturer’s protocols. Control samples were prepared from untreated cells to establish baseline levels of Fe²+ and MDA.
RNA immunoprecipitation assay
The association between IGF2BP2 and SLC7A11 mRNA was examined using the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (cat. No. #17-700; MilliporeSigma). Cell lysates were incubated with magnetic beads conjugated to anti-IGF2BP2 antibody or normal mouse immunoglobulin G (IgG) (negative control) overnight at 4°C. After incubation, the beads were washed and the RNA was eluted from the beads. Following RNA extraction from the immunoprecipitate, SLC7A11 mRNA levels were measured with RT-qPCR as described previously. An IgG control was included to confirm the specificity of the interaction.
mRNA decay assay
To determine mRNA stability, cells were treated with actinomycin D (5 µg/mL, cat. No. #A9415; Sigma-Aldrich) to suppress RNA synthesis. Samples were collected at 0, 2, 4, 8, and 24 h post-treatment. Total RNA was extracted as previously detailed, and SLC7A11 mRNA levels were quantified with RT-qPCR. The decay rates were calculated by normalizing mRNA levels to the amount at time 0. Non-treated cells served as a control for baseline mRNA stability.
Statistical analyses
Data were analyzed using IBM SPSS v. 23.0 (IBM Corp., Armonk, USA). Results were presented as median with interquartile range (IQR) or mean ± standard deviation (SD). The specific statistical test used was determined based on the data distribution and the number of groups compared.
Due to the small sample size (n < 10 per group), non-parametric tests were primarily employed to ensure robust statistical analysis. For comparisons between 2 groups, the Mann–Whitney U test was used. For multiple group comparisons, the Kruskal–Wallis test was applied, followed by Dunn’s post hoc test with Bonferroni correction to identify specific differences between groups.
For experiments involving repeated measurements (e.g., measurements taken at multiple time points for the same group), the Friedman’s test, a nonparametric alternative to repeated-measures analysis of variance (ANOVA), was performed to evaluate overall differences within each group over time. For post hoc pairwise comparisons between time points, Wilcoxon signed-rank tests were applied with Bonferroni correction for multiple comparisons. For comparisons between groups at the final time point, the Kruskal–Wallis test was performed, followed by Dunn’s post hoc test to identify specific between-group differences. A 2-tailed p < 0.05 was considered statistically significant for all analyses.
Results
The IGF2BP2 expression is increased in NSCLC H1299 cells
As demonstrated in a previous study, IGF2BP2 expression is elevated in NSCLC, and its overexpression promotes tumor growth and metastasis.13 Therefore, the differential expression between H1299 cells and HBE cells was first examined. The RT-qPCR findings displayed a remarkable increase in the IGF2BP2 mRNA expression in H1299 cells in comparison with normal HBE cells (U = 0, p = 0.002). The western blot analysis further verified that the IGF2BP2 protein expression was higher in H1299 cells (U = 0, p = 0.002) (Figure 1). These outcomes revealed a marked up-regulation of IGF2BP2 expression in H1299 cells, suggesting that IGF2BP2 had a potential function in H1299 cells.
Effects of IGF2BP2 knockdown and overexpression on H1299 cell viability
We further investigated the role of IGF2BP2 in the H1299 cell by knocking down and overexpressing IGF2BP2. According to the analyses of western blot and RT-qPCR, the IGF2BP2 expression levels were evidently raised in the IGF2BP2 group compared to the vector group, while the si-IGF2BP2 group presented a notable reduction in the IGF2BP2 protein expression relative to the si-NC group (mRNA: H (3, 24) = 19.53, p < 0.001; Protein: H (3, 24) = 19.55, p < 0.001) (Figure 2A,B). These findings indicated that we successfully overexpressed and knocked down IGF2BP2 in H1299 cells. The enhancement or suppression of cell viability in H1299 cells was associated with IGF2BP2 overexpression or knockdown, respectively. As disclosed in the findings from cell viability assay, we observed significant changes in cell viability within each group over time (vector group: χ2 = 12, degrees of freedom (df) = 2, p = 0.002; IGF2BP2 group: χ2 = 10.33, df = 2, p = 0.006; si-NC group: χ2 = 12, df = 2, p = 0.002; si-IGF2BP2 group: χ2 = 12, df = 2, p = 0.002). Post hoc Wilcoxon signed-rank tests revealed that cell viability significantly increased between 24 h and 72 h in each group (p < 0.01). At the final time point (72 h), IGF2BP2 overexpression in H1299 cells significantly increased cell viability, whereas IGF2BP2 knockdown notably reduced it (H (3, 24) = 19.47, p < 0.001) (Figure 2C). These findings suggested that IGF2BP2 promoted the H1299 cell proliferation.
Different effects of IGF2BP2 knockdown and overexpression on ferroptosis in H1299 cells
The accumulation of lipid peroxidation and reactive oxygen species (ROS) leads to ferroptosis, a form of iron-dependent cell death.14 Therefore, further exploration was conducted on the impacts of IGF2BP2 knockdown and overexpression on ferroptosis-related markers in H1299 cells. Figure 3A shows that, as opposed to the vector group, ROS levels were considerably declined in the IGF2BP2 overexpression group, while ROS levels were evidently raised in the IGF2BP2 knockdown group (H(3, 24) = 19.46, p < 0.001). No statistical difference were observed between the vector and siNC groups (p > 0.05) (Figure 3A). Similarly, Fe2+ (H(3, 24) = 19.74, p < 0.001) and MDA (H(3, 24) = 19.56, p < 0.001), indicators of iron accumulation and lipid peroxidation, were significantly lower in the IGF2BP2 group and markedly higher in the si-IGF2BP2 group (Figure 3B,C). ACSL4, a key inducer of ferroptosis,15 and GPX4, an antioxidant enzyme that scavenges ROS and whose inhibition promotes ferroptosis,16 were also upregulated.
IGF2BP2 overexpression considerably increased GPX4 protein expression and decreased ACSL4 levels in H1299 cells. Conversely, IGF2BP2 knockdown markedly decreased the GPX4 protein expression (H (3, 24) = 19.50, p < 0.001) and increased ACSL4 expression (H (3, 24) = 19.61, p < 0.001) (Figure 3D,E). These outcomes indicated that down-regulation of IGF2BP2 could evidently enhance ferroptosis in H1299 cells.
IGF2BP2 modulates SLC7A11 expression through m6A modification
SLC7A11 is a key regulator of ferroptosis; its inhibition induces ferroptosis and suppresses tumor growth.17, 18 We first verified the difference in SLC7A11 expression between normal and cancer cells. In contrast to HBE cells, the SLC7A11 protein expression level was remarkably raised in H1299 cells (U = 0, p = 0.002) (Figure 4A). In H1299 cells, SLC7A11 protein levels were increased in the IGF2BP2 overexpression group and decreased in the si-IGF2BP2 group (H (3, 24) = 19.45, p < 0.001) (Figure 4B). The RNA immunoprecipitation assays confirmed an interaction between IGF2BP2 and SLC7A11 mRNA in H1299 cells (U = 0 p = 0.002), with the m6A modification levels of SLC7A11 mRNA being higher in the IGF2BP2 group and lower in the si-IGF2BP2 group (H (3, 24) = 19.61, p < 0.001) (Figure 4C,D). RNA stability assays demonstrated a time-dependent decrease in SLC7A11 mRNA expression across all groups (χ2 = 18, df = 3, p < 0.001 for all groups). Post hoc Wilcoxon signed-rank tests showed significant differences in SLC7A11 mRNA levels between 0 h and 4 h (p = 0.044), as well as between 0 h and 6 h (p < 0.001), across all groups. At the final time point (6 h), the SLC7A11 mRNA levels were elevated in the IGF2BP2 group and dropped in the si-IGF2BP2 group (H (3, 24) = 18.81, p < 0.001), indicating that IGF2BP2 regulated the stability of SLC7A11 mRNA (Figure 4E). Overall, IGF2BP2 promoted SLC7A11 expression through m6A modification in H1299 cells.
IGF2BP2 regulates cell proliferation and ferroptosis in a SLC7A11-dependent manner
Based on the results presented above, we hypothesized that IGF2BP2 promotes SLC7A11 expression through m6A modification, thereby moderating ferroptosis and cell proliferation. To test this hypothesis, SLC7A11 was overexpressed in H1299 cells following IGF2BP2 knockdown. Initially, we transfected the SLC7A11 plasmid in H1299 cells. Our results suggested that SLC7A11 protein expression was notably raised in H1299 cells transfected with the SLC7A11 plasmid compared to the vector group, indicating successful overexpression (Supplementary Fig. 1).
As illustrated in Figure 5A, a remarkable elevation in cell viability was observed over time in all groups (si-NC + vector group: χ2 = 10.33, df = 2, p = 0.006; si-IGF2BP2 + vector group: χ2 = 12, df = 2, p = 0.002; si-IGF2BP2 + SLC7A11 group: χ2 = 12, df = 2, p = 0.002). Post hoc Wilcoxon signed-rank tests indicated significant differences in cell viability between 24 h and 72 h across all groups (p < 0.01). At the final time point (72 h), cell viability was markedly decreased by IGF2BP2 knockdown as opposed to the siNC + vector group, while the SLC7A11 + si-IGF2BP2 group of H1299 cells exhibited an upward trend in cell viability relative to the si-IGF2BP2 + vector group (H (2, 18) = 13.93, p = 0.001). Correspondingly, the ROS levels were markedly raised in the si-IGF2BP2 + vector group, while a notable decline was disclosed in ROS levels in the si-IGF2BP2 + SLC7A11 group (H (2, 18) = 14.36, p < 0.001) (Figure 5B). Additionally, the si-IGF2BP2 + vector group illustrated a notable elevation in the MDA (H (2, 18) = 14.00, p < 0.001) and Fe2+ (H (2, 18) = 14.78, p < 0.001) levels. However, the si-IGF2BP2 + SLC7A11 group presented a marked downward trend in the MDA and Fe2+ levels (Figure 5C,D).
At the protein level, the si-IGF2BP2 + vector group displayed a remarkable downregulation of SLC7A11 and GPX4 and an upregulation of ACSL4 as opposed to the siNC + vector group. However, these changes were remarkably reversed in the si-IGF2BP2 + SLC7A11 group, with upregulation of SLC7A11 (H (2, 18) = 13.35, p < 0.001) and GPX4 (H (2, 18) = 14.75, p < 0.001) and downregulation of ACSL4 (H (2, 18) = 14.36, p < 0.001) (Figure 5E,F). These outcomes implied that SLC7A11 overexpression reversed the impacts of IGF2BPP2 knockdown on the ferroptosis pathway.
Discussion
A comprehensive study elucidated that IGF2BP2 facilitates a shift in cancer cell metabolism towards glutaminolysis through the post-transcriptional upregulation of SLC7A11, thereby fundamentally contributing to the progression of NSCLC. This metabolic adaptation not only meets the energy and biosynthetic demands of NSCLC cells but also equips them with the ability to counteract oxidative stress, thereby effectively hindering ferroptosis. The elevated levels of IGF2BP2 observed in NSCLC cells are associated with increased cell viability and proliferation, suggesting that IGF2BP2 coordinates the cellular response to the oncogenic stress environments. These outcomes highlight the significance of IGF2BP2 as a therapeutic target and biomarker in NSCLC, which is central to cancer survival, growth and malignant behavior.19, 20, 21
Further study on the role of IGF2BP2 in NSCLC has positioned it as a potential oncogene, with its upregulation linked to enhanced cell proliferation.22 Besides, IGF2BP2 is involved in various cancer-promoting pathways, including cell cycle control,23 evasion of apoptosis24 and metastasis.25 In addition, IGF2BP2 has been reported to activate endothelial cells and promote lung adenocarcinoma angiogenesis and metastasis by single-cell sequencing.26 This aligns with our findings, where we observed that IGF2BP2 promotes NSCLC progression by enhancing cell proliferation and survival, highlighting the role of IGF2BP2 in stabilizing SLC7A11 mRNAs through m6A reading. Furthermore, IGF2BP2 can enhance the colorectal cancer progression by increasing the transferrin receptor expression and promoting iron metabolism.27 Similarly, our study demonstrated that IGF2BP2 stabilized SLC7A11 mRNA, which was vital for maintaining glutathione levels and protecting against ferroptosis in NSCLC.
To fully understand the oncogenic potential of IGF2BP2, future studies should elucidate its interaction network within cancer cells. This includes identifying its mRNA targets and understanding how IGF2BP2 itself is regulated in the cancer setting. Such endeavors will potentially uncover new therapeutic targets and strategies for NSCLC treatment, offering hope for better treatment of this challenging disease. The regulatory role of IGF2BP2 extends beyond metabolic pathways to include the stabilization of oncogenic mRNAs, such as SLC7A11, which is essential for maintaining redox homeostasis and protecting against ferroptosis.4 In this study, a significant mechanism in NSCLC pathology was revealed. IGF2BP2 stabilizes SLC7A11 mRNA, which is the core of maintaining cellular redox balance. SLC7A11 ensures the uptake of cystine, which is necessary for glutathione synthesis, enabling cells to withstand oxidative stress and avoid ferroptosis. The effect of IGF2BP2 on SLC7A11 marks a protective mechanism for NSCLC cells, allowing them to thrive in a pro-oxidant tumor microenvironment. The relationship between IGF2BP2 and SLC7A11 suggests that cellular metabolism is closely associated with susceptibility to ferroptosis, potentially reshaping our approach to cancer treatment.
Although this study focused on mRNA stabilization, IGF2BP2 might regulate SLC7A11 through other mechanisms, such as post-translational modifications (like phosphorylation), miRNA interaction and protein-protein interaction.28, 29, 30 In the future, several approaches including phosphoproteomic analysis, miRNA profiling and protein interaction analysis could be employed to further explore these potential mechanisms to comprehensively understand the IGF2BP2’s role in regulating SLC7A11.
As is reported, metabolic reprogramming in NSCLC, particularly the shift towards glutaminolysis,31 is not merely a response to energy demands but a strategic adaptation that confers survival advantages.32 Through this pathway, NSCLC cells acquire antioxidant capacity by generating nicotinamide adenine dinucleotide phosphate (NADPH), which supports the regeneration of reduced glutathione. The role of IGF2BP2 in this metabolic shift not only supports rapid cell proliferation but also contributes to the cells against ferroptosis.33 Given this adaptability, targeting metabolic reliance in NSCLC may prove effective, especially when paired with therapies that induce ferroptosis.34, 35 The therapeutic potential of interfering with the IGF2BP2-SLC7A11 interaction and glutaminolysis is substantial. The development of small molecule inhibitors to disrupt this interaction could directly counter the ferroptosis resistance in NSCLC cells. Additionally, targeting glutaminolysis can deplete the metabolic resources of NSCLC cells, making them susceptible to ferroptosis. Such strategies, particularly when combined with existing treatments, can revolutionize NSCLC therapy. The current challenge is to translate these findings into clinical applications, which will involve the development and rigorous testing of targeted pharmacological agents in preclinical and clinical settings. Our findings support a precision medicine approach, suggesting that patient-specific therapies based on the expression of IGF2BP2 and SLC7A11 could enhance treatment outcomes. This study lays a solid foundation for future efforts to conquer NSCLC, highlighting the IGF2BP2–SLC7A11 axis standing as a promising target for novel cancer therapies.
Importantly, this study uncovers a previously unreported regulatory axis where IGF2BP2 stabilizes SLC7A11 mRNA through m6A reading, providing a novel understanding of how IGF2BP2 contributes to metabolic adaptation and ferroptosis resistance in NSCLC. This novel mechanism highlights IGF2BP2 as a potential therapeutic target, opening new avenues for developing targeted treatments to improve clinical outcomes in NSCLC.
Limitations
However, certain limitations must be acknowledged. First, we relied on the MTT assay to assess cell viability. However, although this method is efficient and widely accepted, it does not reflect other aspects of cell behavior such as proliferation rate, migration and invasion capabilities. Therefore, further studies incorporating comprehensive assays such as colony formation, migration and invasion are necessary to thoroughly evaluate malignant behavior. Second, this study was conducted primarily in vitro. The use of cell lines may not fully represent the complexity and heterogeneity of tumor behavior in vivo. Moreover, the sample size used in our study was relatively small, which may limit the statistical power and generalizability of the results. Future studies with larger sample sizes will help confirm the robustness of these findings and provide more reliable statistical analysis. Additionally, our study merely offers preliminary data suggesting potential ferroptotic activity and cell proliferation, necessitating additional specific assays to comprehensively confirm ferroptosis. Furthermore, we need to consider the potential compensatory mechanisms that could arise from the inhibition of the IGF2BP2-SLC7A11 axis.12 Ultimately, although IGF2BP2 has been shown to regulate the stability of SLC7A11 and affect cell viability and ferroptosis, the precise molecular mechanisms and downstream signaling pathways have only been partially elucidated. Comprehensive proteomic and metabolic studies may reveal additional layers of regulation and interaction that contribute to the observed phenotypes. These limitations highlight the importance of future studies with multidimensional approaches, integrating genomic, proteomic and metabolomic data to fully elucidate the role of IGF2BP2 in NSCLC.
Conclusions
IGF2BP2 plays an essential role in NSCLC by modulating m6A modification of SLC7A11 mRNA and enhancing its stability, thereby inhibiting ferroptosis. These findings highlight the therapeutic potential of targeting the IGF2BP2-SLC7A11 axis. Future research should focus on exploring the broader clinical implications, such as the development of small molecule inhibitors or other therapeutic strategies that target this pathway. Additionally, investigating the role of IGF2BP2 in other types of cancers may reveal new insights into its role in cancer progression and ferroptosis regulation, potentially leading to more effective, personalized treatments.
Supplementary data
The supplementary materials are available at https://doi.org/10.5281/zenodo.15074419. The package includes the following files:
Supplementary Fig. 1. Successful overexpression of SLC7A11 in H1299 cells.
Data availability
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
Consent for publication
Not applicable.
Use of AI and AI-assisted technologies
Not applicable.

















.jpg)
.jpg)
.jpg)
.jpg)
.jpg)