Abstract
Background. With the increasing number of older pregnant women and environmental pollution, the incidence of congenital malformations increases every year. Prenatal diagnosis is an effective means of identifying congenital malformations.
Objectives. To evaluate the clinical utility of single nucleotide polymorphism (SNP) microarray analysis during prenatal evaluations.
Materials and methods. To assess the similarities and differences between the 2 approaches, 425 pregnant women were selected to undergo prenatal gene SNP microarray analysis and karyotype analysis during prenatal evaluation between January 2020 and August 2021.
Results. The success rate of SNP microarray analysis was 100%, which was statistically different from that of karyotype analysis (92%, Fisher’s exact test, p < 0.001). The positive rate of SNP detection was 10.4% higher than karyotype analysis, which was 6.6% (Pearson’s χ2 test, χ2 = 3.89, degrees of freedom (df) = 1, p = 0.049). Karyotype analysis detected 28 cases of aneuploidy; SNPs could not only detect these results of karyotype analysis, but 16 cases of copy number variations (CNV) with obvious pathogenicity, including duplications/deletions, chimerism and loss of heterozygosity (LOH).
Conclusions. Single nucleotide polymorphism microarray analysis technology is an important method used in prenatal genetic evaluations, which can find fetal genetic etiologies, correctly evaluate the fetal prognosis during prenatal clinical examination, and provide a more objective basis for whether to continue the pregnancy.
Key words: karyotype analysis, prenatal diagnosis, SNP microarray analysis
Background
With the increasing number of older pregnant women and environmental pollution, the incidence of congenital malformations increases every year. Prenatal diagnosis is an effective means to diagnose congenital malformations.1 Karyotype analysis is currently the gold standard for the prenatal diagnosis of genetic abnormalities, but it also has drawbacks, including the need for cell culture, long reporting cycles, and an increased risk of sample contamination that can lead to experimental failure.2 Karyotype analysis can distinguish only fragments larger than 10 megabase (Mb) pairs, and often misses some disease-causing genes smaller than 10 Mb. Microdeletion and microreplication syndrome3 is caused by the duplication or deletion of chromosomal segments smaller than 5–10 Mb and is associated with a wide range of malformations and dysgnosia. Microreplication and microdeletion of genomic segments (also known as copy number variation (CNV)) are associated with 6–15% of genetic diseases. Detection of CNVs requires high-resolution technology for which traditional karyotype analysis is ineffective.4 Because of its rapid, high-throughput and high-resolution ability, chromosomal microarray analysis (CMA) has been accepted as a powerful tool for the diagnosis of developmental retardation, intellectual disability, autism, and multiple congenital anomalies.5, 6, 7, 8 Microdeletions and microreplications as small as 50–100 kilobase (kb) pairs can be detected using CMA across the entire genome.9 Chromosomal microarray analysis is currently classified into 2 categories: array-based comparative genomic hybridization (aCGH) and single nucleotide polymorphism (SNP) array. The aCGH demonstrates the relative amounts of DNA from various areas of the genome by comparing the test DNA sample with a normal reference DNA sample and cannot detect triploidy, while SNP arrays can identify triploidy as well as loss of heterozygosity (LOH) by hybridizing the test sample to the array platform and analyzing the signal intensity of SNP probes.10, 11 The SNP array analysis technology is a new molecular method for karyotype analysis with high resolution, sensitivity, throughput, and accuracy, capable of detecting and analyzing submicroscopic changes in ploidy.12 The SNP array analysis is effective in the prenatal diagnosis of genetic causes of fetal nasal bone deletion.13 The SNP array technology has been increasingly applied in prenatal clinical genetic evaluations and has become a recommended genetic testing method for the prenatal diagnosis of fetal growth retardation, mental retardation, autism, and organ malformations, especially when an ultrasound shows fetal abnormalities and a normal karyotype.14
Objectives
To evaluate the clinical utility of SNP arrays in prenatal evaluations.
Objects and methods
Research objects
Prenatal SNP array analysis and karyotype analysis were performed on 425 pregnant women aged 19–45 years old between 16 and 31 weeks gestation at our antenatal diagnostic center between January 2020 and August 2021. The main indications of prenatal diagnosis were abnormal prenatal ultrasonography, chromosomal abnormalities of one spouse, advanced maternal age, fetal structural abnormalities, and noninvasive genetic abnormalities. All pregnant women enrolled in the study received technical counseling and signed informed consent forms for the SNP array and conventional karyotypes prior to the study.
Materials and methods
Chromosome karyotype analysis
After samples were centrifuged at 2,000 rpm for 10 min, the supernatant was discarded and the samples were incubated for 8–9 days. Each sample was counted and analyzed for 20–30 mitotic periods, and 5 radionuclide images were observed, analyzed and photographed. If any abnormality was detected, the number of karyotypes increased. Using the International Nomenclature of Human Cytogenetics (ISCN, 2005),15 the aberrant karyotype was described.
Single nucleotide polymorphism array analysis
Genomic DNA was extracted from fetal umbilical cord blood and amniotic fluid that had never been cultured or previously cultured using a Qiagen kit (Qiagen, Hilden, Germany). The SNP analysis was performed using a genome-wide Affymetrix CytoScan 750 K array (Affymetrix Inc., Santa Clara, USA) containing 500,000 probes for CNV and 200,000 probes for SNPs spread throughout the whole human genome. Data were analyzed using the human genome version of GRCh37 (hg19) and Chromosome Analysis Kit software (Affymetrix). Gains or losses ≥400 kb and LOH ≥ 10 Mb were the most common recommendations. Genome-wide identification of uniparental disomy (UPD) in the offspring-parents triad can be confirmed as having a maternal or paternal origin using the UPDtool.16 All CNVs found were compared using the following databases: Database of Genomic Variants (DGV, http://projects.tcag.ca/variation), Database of Chromosomal Imbalance and Phenotype in Humans using Ensemble Resources (DECIPHER) database (https://www.deciphergenomics.org), International Standards for Cytogenomic Arrays, and Online Mendelian Inheritance in Man (OMIM) database (http://www.omim.org).17
Statistical analyses
For qualitative data, the assumption of expected abundance for the χ2 test was checked. If the assumption was met, Pearson’s χ2 test of independence was used to analyze the differences between groups. If the assumption was not met, Fisher’s exact test for 2×2 tables was used. The analysis was performed using GraphPad Prism v. 9.0 (GraphPad Software, San Diego, USA).
Results
General conditions of study objects
Age composition of objects
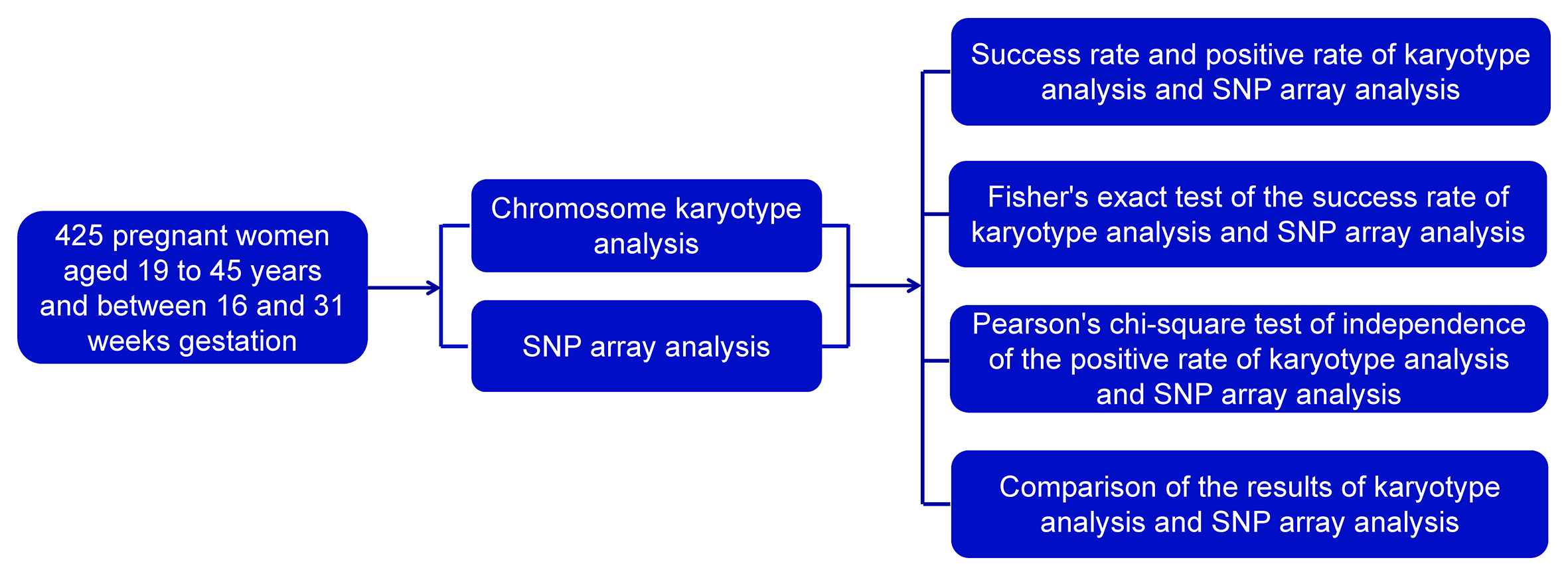
There were 425 pregnant women aged 18–45 years old. The distribution of cases by age group is illustrated in Figure 1. The 18–23 age group had 27 cases, the 24–29 age group had 105 cases, the 30–34 age group had 133 cases, the 35–39 age group had 122 cases, and the 40–45 age group had 38 cases.
Gestational weeks of study participants
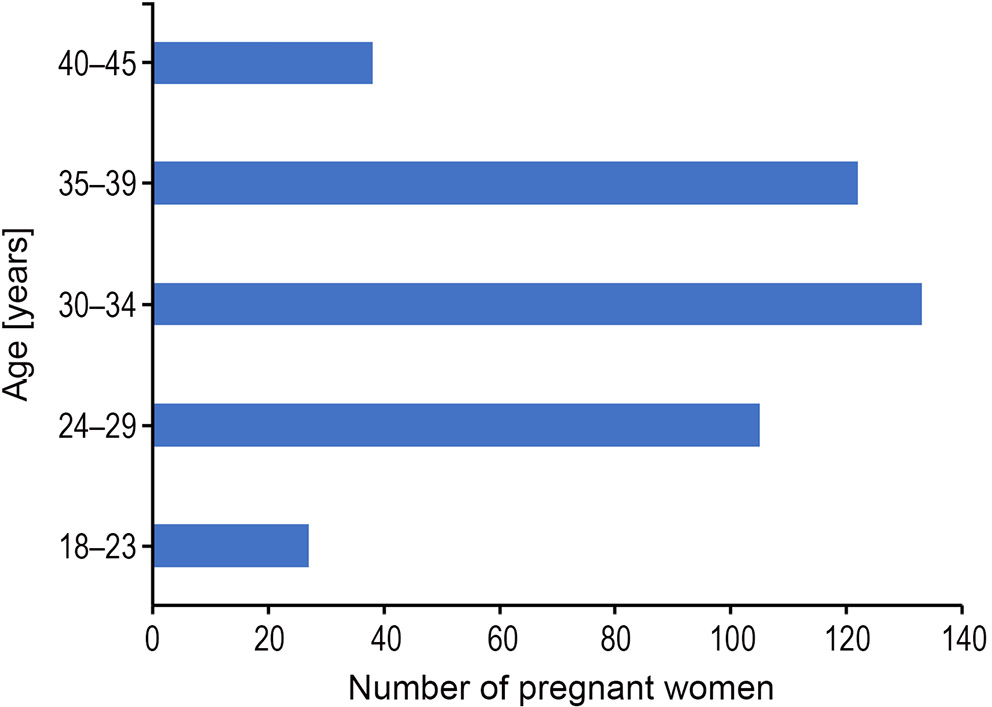
The prenatal diagnosis of 425 pregnant women was between 16 and 31 weeks, as shown in Figure 2. There were 7 cases in the 16+ weeks group, 65 cases in the 17+ weeks group, 89 cases in the 18+ weeks group, 70 cases in the 19+ weeks group, 67 cases in the 20+ weeks group, 28 cases in the 21+ weeks group, 20 cases in the 22+ weeks group, 32 cases in the 23+ weeks group, 11 cases in the 24+ weeks group, 11 cases in the 25+ weeks group, 5 cases in the 26+ weeks group, 2 cases in the 27+ weeks group, 2 cases in the 28+ weeks group, 4 cases in the 29+ weeks group, 10 cases in the 30+ weeks group, and 2 cases in the 31+ weeks group.
Sample type of the study objects
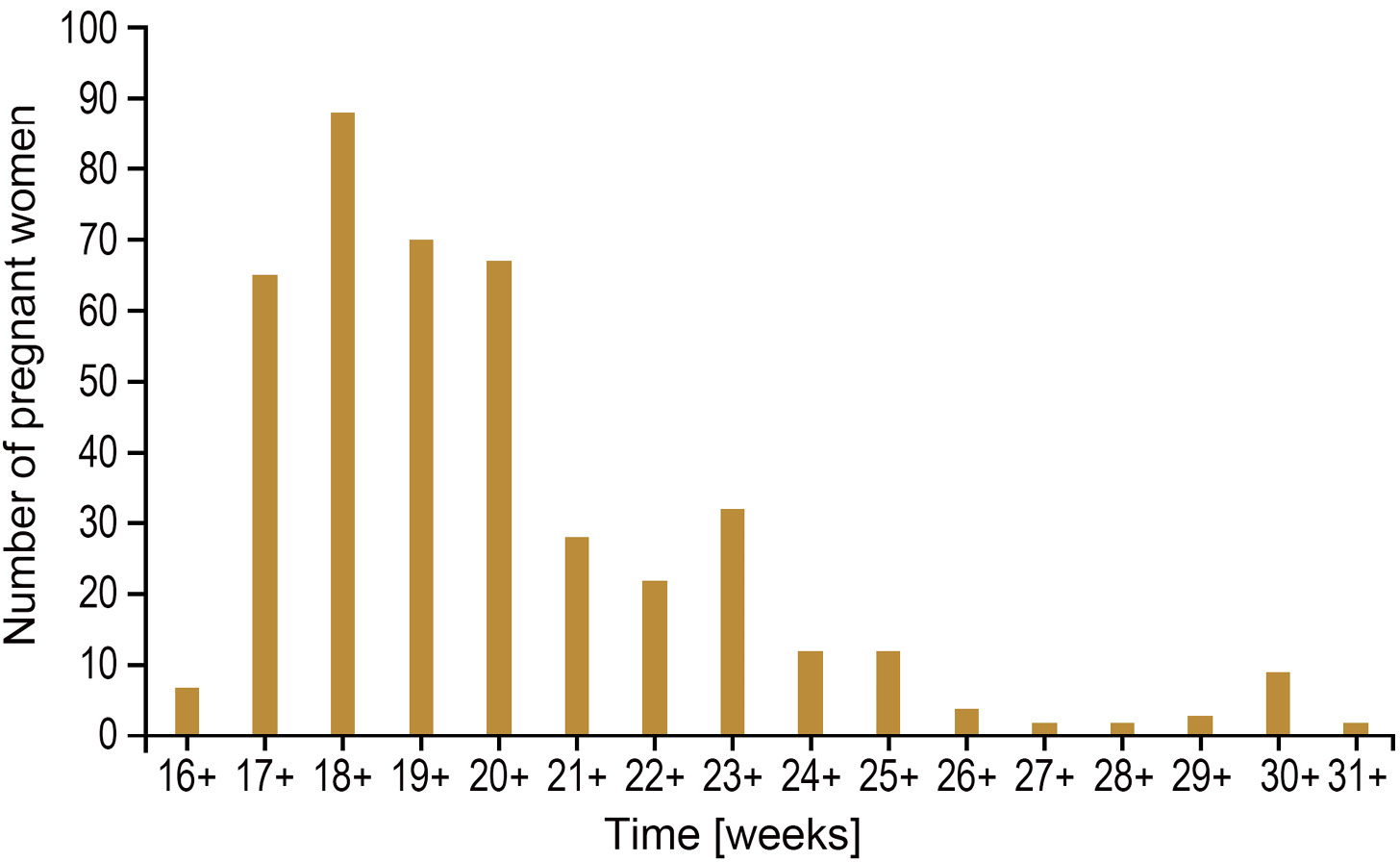
Among the 425 samples, 363 were amniotic fluid, accounting for 85.41% of the total sample size, 30 were miscarriage villi, accounting for 7.06% of the total sample size, and 32 were umbilical cord blood, accounting for 7.53% of the total sample size, as shown in Figure 3.
Success rate and positive rate of karyotype analysis and SNP array analysis
There were 425 samples, with 391 successful and 34 failed cases (a success rate of 92%). Karyotype analysis identified 28 cases of aberrant karyotypes among the 425 samples, with a positive rate of 6.6%, as shown in Table 1.
In addition, the success rate of SNP array analysis was 100%. Compared with karyotype analysis, the success rate was significantly improved (Table 2, Fisher’s exact test, p < 0.001). Forty-four cases showed positive results, with a positive rate of 10.4%, which was statistically significant compared to karyotype analysis (Table 3, Pearson’s χ2 test, χ2 = 3.89, degrees of freedom (df) = 1, p = 0.049).
Results of karyotype analysis and SNP array analysis
Among the 28 cases with abnormal karyotypes detected with karyotype analysis, there were 12 cases with trisomy 21, 4 cases with trisomy 18, 2 cases with trisomy 16, 1 case with trisomy 13, 5 cases with 45 X, 1 case with 46 XXX, and 3 cases with 47 XXY. In addition to the abnormal cases, 16 cases with normal karyotypes but abnormal SNP array analysis were also detected, including 8 cases of duplication/deletion, 3 cases of chimera and 5 cases of LOH. Table 4 summarizes the outcomes.
Results of normal karyotype analysis and abnormal SNP array analysis
Sixteen CNVs with normal karyotype were found to be abnormal in the SNP array analysis results, with a fragment length of 349 kb–14.59 Mb, which was related to microdeletion, microreplication, chimera, and LOH syndrome, as shown in Table 5.
Discussion
Although traditional chromosome karyotyping can identify polyploidy, aneuploidy, translocation, inversion, chimerism, duplication, and deletions larger than 10 Mb, this technique has various limitations, including time-consuming cell cultures, low resolution, high labor demand, and an inability to identify CNVs smaller than 10 Mb.18, 19, 20 Therefore, there is an urgent need for a new testing technique to be used in prenatal evaluations.
Karyotype analysis and CMA are 2 prenatal diagnostic methods that have been widely used over the recent years.21, 22, 23 Chromosomal microarray analysis is a high-resolution technology for whole-genome analysis, detecting micro-deletions and micro-duplications, which are not routinely detected via karyotyping. Chromosomal microarray analysis can be separated into aCGH and SNP array. The SNP array can detect regions of homozygosity, triploidy, and maternal cell contamination, which the aCGH cannot. The SNP array analysis has been widely used in the field of prenatal genetics and has shown tremendous advancement and superiority as a result of scientific and technological developments.24 Compared to conventional karyotyping, the resolution of SNP array analysis is much greater, allowing the identification of submicroscopic imbalances such as UPDs and LOHs.25 The value of the additional information provided by SNP array analysis in prenatal diagnosis is evident in reducing the prevalence of fetuses at increased risk of chromosomal imbalances that are undetected or may have genetic disorders.26 The SNP array analysis can influence pregnancy care and outcomes by offering more precise prognostic information than karyotype analysis.27
Compared with traditional karyotypes, the time for SNP array analysis is much shorter, and the time for patients to wait for the genetic results is also significantly shortened.3 In this study, 425 amniotic fluid, miscarriage villi and umbilical cord blood samples were subjected to traditional cell culture, and karyotype analysis generally takes 12–15 days, with some samples taking up to 20 days due to low cell activity and slow growth.28 In contrast, the detection period for SNP array analysis is only 3–4 days. Prenatal evaluations are an important means to judge the health state of the fetus. The rapid results of genetics can reduce the waiting time of pregnant women and greatly reduce the psychological burden.29
In addition, SNP array analysis technology has the advantages of simple operation and speed, while traditional karyotype analysis technology requires a high level of operation skills, strict aseptic operation of cell cultures, and a tedious and time-consuming chromosome preparation process. The SNP does not require a complex chromosome preparation process or strict aseptic operation, and it only utilizes a modest amount of DNA to identify the whole genome’s changes in copy number.30 In this study, 425 samples were cultured under strict aseptic conditions, and 391 samples were successfully cultured with a success rate of 92%. Some samples failed because cells did not adhere to cell walls after culture or cell cloning was stunted, and karyotype isolates were not collected for analysis. This limitation was overcome by the SNP array analysis technology, which obtained results for all 34 samples that failed in culture with a 100% success rate, significantly higher than the karyotyping technique.
The SNP array analysis technology can provide a higher resolution.31 The number of chromosome bands prepared by traditional karyotype technology is between 300 and 420, and the resolution is low. Theoretically, only 5–10 Mb chromosome structural abnormalities can be detected.32 The SNP array analysis technology has high throughput and resolution, enabling the detection of genome-wide CNVs at one time, as well as detecting single gene diploid and small chromosome variants that cannot be identified by karyotype analysis.33 In addition, SNP probes also provide SNP typing information, which can detect heterozygous loss and can be used for clinical detection of some residual and imprinted genetic diseases. In this study, among the cases with normal karyotypes, 16 CNVs with specific pathogeneses were detected using SNP array analysis, and the length ranged from 349 kb to 14.59 Mb, which involved the microdeletion of a 367 kb fragment in the p12.2 region of chromosome 10, the microreplication of a 588 kb fragment in the q26 region of chromosome 6 and the ROH of about 13.6 Mb in the q15q21.3 region of chromosome 5.
The inability of SNP array analysis to recognize balanced chromosomal structural abnormalities, such as balanced translocations and reversals,34 prevents them from completely replacing karyotype analysis, despite its many benefits.35 Although most of these abnormalities are inherited from the parents and do not necessarily affect the phenotype of the fetus, they may still cause spontaneous abortion throughout the reproductive process or cause chromosomal imbalances.36 A small number of mosaic chromosomal defects may also be undetectable using SNP array analysis. Given that SNP array and karyotype analyses are not substitutable for each other, the combination of the 2 can provide us with more genetic information to avoid a missed diagnosis and provide more basis for genetic counseling.37
Limitations
A larger number of participants is suggested to reduce result bias. Moreover, to popularize and generalize SNP array analysis technology, more studies are needed.
Conclusions
The SNP array analysis is an important tool for screening chromosome deletions and replications, especially in fetuses with normal karyotypes. With technical improvements and data accumulation, SNP array analysis will play an important role in prenatal diagnosis.
Ethics statement
The present study was approved by the Ethics Committee of Maternity and Child Care Hospital of Huaihua (approval No. 20200925CZ) and written informed consent was provided by all patients prior to the study. All procedures were performed in accordance with the ethical standards of the Institutional Review Board and The Declaration of Helsinki, and its later amendments and comparable ethical standards.
Data availability
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
Consent for publication
Not applicable.
Use of AI and AI-assisted technologies
Not applicable.