Abstract
Background. Recurrent miscarriage (RM) affects 1–2% of couples. Maternally expressed gene 3 (MEG3) is aberrantly expressed in RM patients.
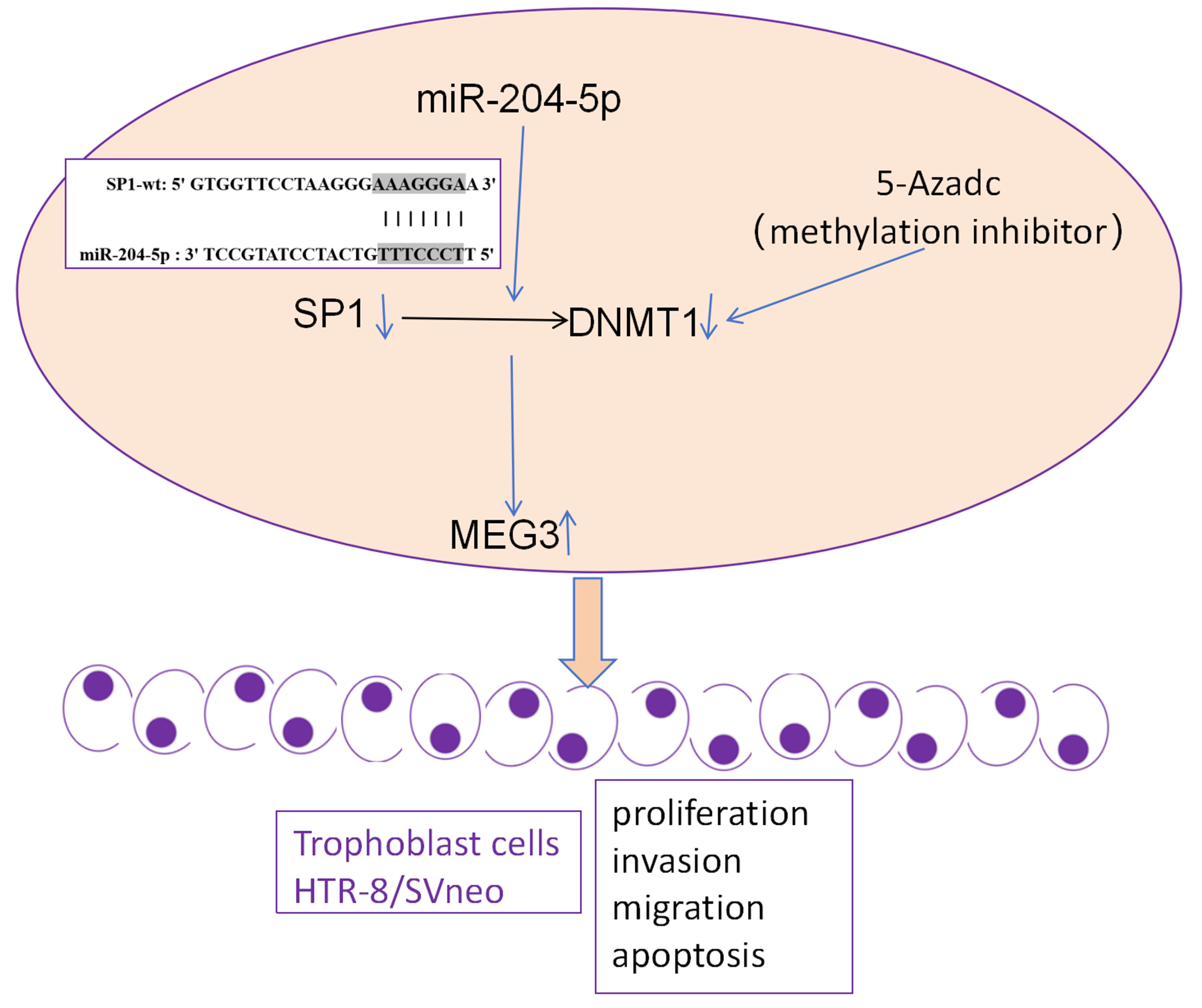
Objectives. To investigate a novel regulatory mechanism, we examined the miR-204-5p/Specificity protein 1 (SP1)/DNA methyltransferase 1 (DNMT1)/MEG3 axis in the trophoblast cell line HTR-8/SVneo.
Materials and methods. Human trophoblast cell line HTR-8/SVneo was used and cells were transfected with siRNA targeting SP1, miR-204-5p mimics, pcDNA3.1-DNMT1, or their negative controls (NCs). The methylation inhibitor, 5-azadC, was used to treat the cells transfected with pcDNA3.1-SP1. The reverse transcription quantitative polymerase chain reaction (RT-qPCR) method was used to examine the relative RNA levels of SP1, DNMT1 and MEG3. Western blot assay was performed to measure the protein levels of SP1 and DNMT1. The dual-luciferase reporter gene assay was used to validate the miR-204-5p bindings to SP1. Functional assays were utilized to assess cell apoptosis, colony formation, migration, and invasion.
Results. SP1 knockdown inhibited DNMT1 and increased MEG3 expression. The expression of MEG3 was enhanced by methylation inhibition through 5-azadC, but SP1 upregulation reversed this effect. SP1 knockdown increased apoptosis and decreased migration and invasion, which was reversed by DNMT1 overexpression. SP1 was targeted and inhibited by miR-204-5p. miR-204-5p also inhibited DNMT1, and enhanced the expression of MEG3. miR-204-5p inhibited cell proliferation, migration and invasion, and promoted apoptosis. Overexpression of SP1 partially reversed these effects by activating DNMT1 and inhibiting MEG3.
Conclusions. miR-204-5p promoted MEG3 expression in trophoblast cells via SP1-mediated DNMT1 inhibition, leading to reduced cell migration, proliferation and invasion, as well as increased apoptosis. This study reveals a novel regulatory axis in trophoblast cells, providing insights into potential regulatory mechanisms in RM.
Key words: MEG3, SP1, DNMT1, recurrent miscarriage, trophoblast cells
Background
Recurrent miscarriage (RM) affects 1–2% of women attempting to conceive, imposing both emotional and clinical burden.1 Despite extensive research, the underlying causes of RM remain incompletely understood, often involving a complex interplay of genetic, epigenetic and environmental factors, including endocrine disruptors such as mancozeb.2, 3, 4, 5, 6, 7
Recent advances in molecular biology have revealed the crucial role of extravillous trophoblast cells in maintaining a successful pregnancy by mediating implantation and placental development.8 Previous research has also shown that severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) affects female reproductive health by targeting trophoblasts.9 Dysfunction of trophoblasts is a well-established contributor to pregnancy loss.
Previously, specificity protein 1 (SP1), also known as a transcriptional factor SP1, was found to be reduced in the chorionic villus tissues in the placentas from RM patients, and the downregulation of SP1 by upregulation of miR-4497 could induce the apoptosis of trophoblast cells, which is associated with RM occurrences.10, 11 SP1 regulates genes essential for cellular growth and differentiation, while DNMT1 is correlated with DNA methylation patterns, and is of significance in maintaining genomic stability and gene expression.12, 13 Similar to miR-4497, miR-204-5p was also involved in studies focusing on unexplained recurrent spontaneous abortion.14 miR-204-5p plays a role in placental development and function. For instance, miR-204-5p has been revealed to modulate the trophoblast cell proliferation and invasion, which are essential for proper placental development.15 Dysregulation of miR-204-5p was revealed to be associated with pregnancy-related disorders, including preeclampsia (PE).16, 17 miR-204-5p expression was downregulated in PE and the upregulation of miR-204-5p was related to the decreased migration of trophoblast cells.17
Long non-coding RNA (lncRNA), such as maternally expressed gene 3 (MEG3), is a known regulator in tumor suppression and regulation of cell growth and death.18 Studies have unveiled that MEG3 expression is downregulated in blood samples, embryonic villus and chorionic villus tissues from RM patients, indicating its potential role in the pathogenesis of RM.19, 20, 21 Further, MEG3 was validated to stimulate the invasion and proliferation of trophoblast cells.20, 21 Similarly, in PE, downregulation of MEG3 was discovered to increase apoptosis of trophoblast cells and inhibit cell invasion.22 Previously, DNMT1 was discovered to enhance methylation of MEG3 promoter and decrease MEG3 expression, thereby mediating the disease progression in breast cancer,23 anemia,24 diabetic retinopathy,25 etc. However, such interaction between DNMT1 and MEG3 has not been validated in trophoblast cells.
In this study, we hypothesized that miR-204-5p/SP1/DNMT1 axis plays a pivotal role in modulating trophoblast cell invasion, proliferation and apoptosis, processes essential for placental development and successful pregnancy, through MEG3 modulation via epigenetic modifications.
Objectives
Our research aims to elucidate a novel modulatory mechanism of miR-204-5p in trophoblast cells via SP1/DNMT1/MEG3 pathway and its impact on cell invasion, proliferation and apoptosis.
Materials and methods
Culture of cells
HTR-8/SVneo cells, derived from human first-trimester chorionic villi, were obtained from Procell (Wuhan, China). The cells were cultured in Roswell Park Memorial Institute Medium (RPMI) 1640 medium supplemented with 10% fetal bovine serum (FBS; Gibco, Waltham, USA) at 37°C in a humidified atmosphere containing 5% CO₂.
Cell transfection and treatment
Cells were reseeded in 6-well plates at a density of 5 × 10⁵ cells per well and incubated overnight. On the 2nd day, cells were transfected with specific siRNAs, overexpressed plasmids, miRNA mimics, or their negative controls (NC) (GenePharma, Tianjin, China), using Lipofectamine 2000 kit (Invitrogen, Waltham, USA). Following the protocols, cells were transfected with 50 nM siRNA targeting SP1 (si-SP1) or negative control siRNA (si-NC) (GenePharma). For miRNA overexpression, cells were transfected with 50 nM miR-204-5p mimics (miR-204-5p), mimics negative control (NC mimics). For overexpression of SP1 or DNMT1, cells were collected for transfection, with 2 μg of pcDNA3.1 empty vector (Ctrl), pcDNA3.1-SP1 (SP1) or pcDNA3.1-DNMT1 (DNMT1) (GenePharma). In addition, the methylation inhibitor, 5-azadC (1 μM; Aladdin, Tianjin, China) was used to treat the cells for methylation inhibition.
RNA isolation and reverse transcription quantitative polymerase chain reaction method
RNA was extracted from all cell groups using TRIzol Kit (Beyotime, Beijing, China). Complementary DNA (cDNA) was synthesized using a PrimeScript RT reagent Kit (TaKaRa, Tokyo, Japan). The reverse transcription quantitative polymerase chain reaction (RT-qPCR) assay was performed using the SYBR Taq II Kit (TaKaRa) on Step OnePlus RT-PCR (Applied Biosystems, Waltham, USA). The specific primers used for RT-qPCR are listed below:
miR-204-5p:
Forward(F) 5’-TGCGGTTCCCTTTGTCATCCTATG-3’;
Reverse (R) 5’-GTCGTATCCAGTGCAGGGTCCGAGGTGCACTGGATACGACAGGCATAG-3’.
U6:
F 5’-CTCGCTTCGGCAGCACA-3’,
R 5’-AACGCTTCACGAATTTGCGT-3’.
SP1:
F 5′-CTGGTCCCATCATCATCCGG-3′,
R 5′-TGTTTGGGCTTGTGGGTTCT-3′.
DNMT1:
F 5′-TGGTGAAGACGCCAGTGGA-3′,
R 5′-CGTGGCTGTGGAGGGATTTCG-3′.
MEG3:
F 5′-CGGCTGAAGAACTGCGGATGG-3′,
R 5′-CGTGGCTGTGGAGGGATTTCG-3′.
GAPDH:
F 5’-GCACCGTCAAGGCTGAGAAC-3’,
R 5’- TGGTGAAGACGCCAGTGGA -3’.
The expression levels were analyzed in Microsoft Excel 2013 software (Microsoft Corp., Redmond, USA) using the 2–ΔΔCt method, with U6 (for miR-204-5p) or GAPDH (for the rest) as the house keeping reference.
Western blot analysis
Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer with phenylmethylsulfonyl fluoride (PMSF) (Beyotime). Proteins were separated by using the sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) methods, and transferred to polyvinylidene difluoride (PVDF) (Millipore, St. Louis, USA). The membranes were then blocked in 5% skim milk for 1 h at room temperature and then incubated overnight at 4°C with the following primary antibodies: SP1 (1:1000, #5931; Cell Signaling Technology (CST), Danvers, USA), DNMT1 (1:1000, #5032; CST) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (1:10,000, AB2000; Abways, Tianjin, China). Membranes were incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies (1:20,000, AB0101; Abways) for 1 h at room temperature. Immunoblots were then visualized using an enhanced chemiluminescence (ECL) kit, and images were captured using the SCG-W3000 imaging system (ServiceBio, Wuhan, China).
Dual-luciferase reporter assays
The binding sites were predicted using StarBase database (https://rnasysu.com/encori). The pmirGLO-SP1-3′UTR-wild type (SP1-UTR-wt) and mutant type (SP1-UTR-mt) were constructed using the pmirGLO vector (Promega, Madison, USA). Cells were co-transfected with 50 ng of SP1-UTR-wt or SP1-UTR-mt and 50 nM miR-204-5p mimics/inhibitors, or respective controls (GenePharma). After 48 h, luciferase activity was measured on a multi-functional microplate after using the Dual-Luciferase Reporter Assay kit (Beyotime).
Flow cytometry for apoptosis analysis
Apoptosis was assessed using Cell Apoptosis Kit (Bioss, Beijing, China). Transfected cells were collected, washed with precooled phosphate-buffered saline (PBS) and suspended using binding buffer. As per the manufacturer’s protocol, cells were stained in Annexin V-FITC and propidium iodide (PI) for 15 min at room temperature in dark. Cell apoptosis was analyzed with flow cytometry (BD FACSCanto II; BD Biosciences, Franklin Lakes, USA).
Colony formation assay
Cells after transfection were collected and reseeded in 6-well plates with a density of 600 cells per well. Cells were observed every day till the formation of colonies. Cells were fixed using 4% paraformaldehyde for 20 min and then stained with 0.1% crystal violet for 30 min in dark. Colonies with more than 50 cells were counted in each group.
Transwell assays
The transwell chambers with 8-μm pores were obtained from Jet Biotech (Guangzhou, China). For migration assays, cells were washed in PBS twice and seeded in the upper chamber with serum-free medium at a density of 5 × 10⁴ per well, while culture medium with 15% FBS was added in the lower chamber. For invasion assays, the upper chambers were pre-coated with Matrigel (Corning Company, Corning, USA). Cells were observed under an optical microscope (model Ts2; Nikon Corp., Tokyo, Japan) every 6 h. After 36 h, cells that had migrated or invaded to the lower surface were washed using PBS twice gently and fixed using 4% paraformaldehyde for 20 min, and then stained in crystal violet for 30 min. Cells were then observed and counted under an optical microscope (model Ts2; Nikon Corp.) from 5 different fields.
Statistical analyses
Due to the small sample size, non-normal distribution was assumed. The statistical significance of difference was determined with the nonparametric Kruskal–Wallis test with Dunn’s post hoc test, and Mann–Whitney U test for comparison within 2 groups using GraphPad v. 9.0 (GraphPad Software, San Diego, USA). Bonferroni correction was performed for multiple testing. The results from statistical analysis in this study were included in Supplementary Tables 1 and 2. The figures were generated based on statistical data and cell images from experiments mentioned above using GraphPad. Data in replicates are presented in figures as scatter dots with the median and range lines. The p-values less than 0.05 were considered statistically significant.
Results
Downregulation of SP1 promoted MEG3 through methylation inhibition in trophoblast cells
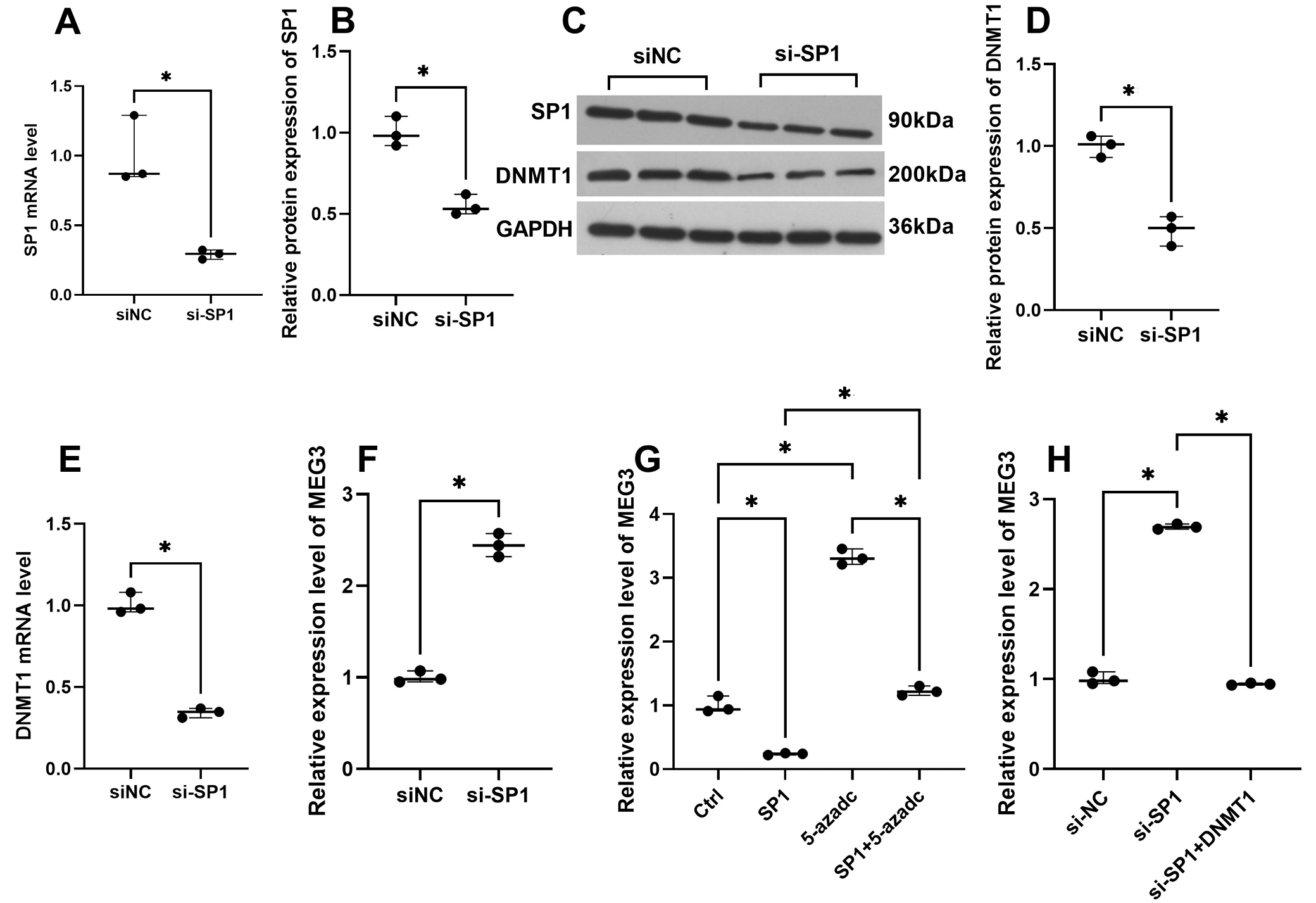
SP1 mRNA and protein levels were inhibited in HTR-SV/neo cells transfected with si-SP1 (Figure 1A–C). Furthermore, SP1 knockdown in cells inhibited both DNMT1 mRNA and protein levels (Figure 1C–E). Additionally, SP1 knockdown upregulated MEG3 expression, an effect that was reversed by DNMT1 overexpression (Figure 1F,H). A methylation inhibitor, 5-azadC, promoted the MEG3 expression and overexpression of SP1 could partly reverse the effect of 5-azadC (Figure 1G). The findings suggest that SP1 knockdown enhanced MEG3 by DNMT1-mediated methylation in trophoblast cells.
SP1 knockdown decreased trophoblast cell invasion, proliferation and migration and increased apoptosis through DNMT1 modulation
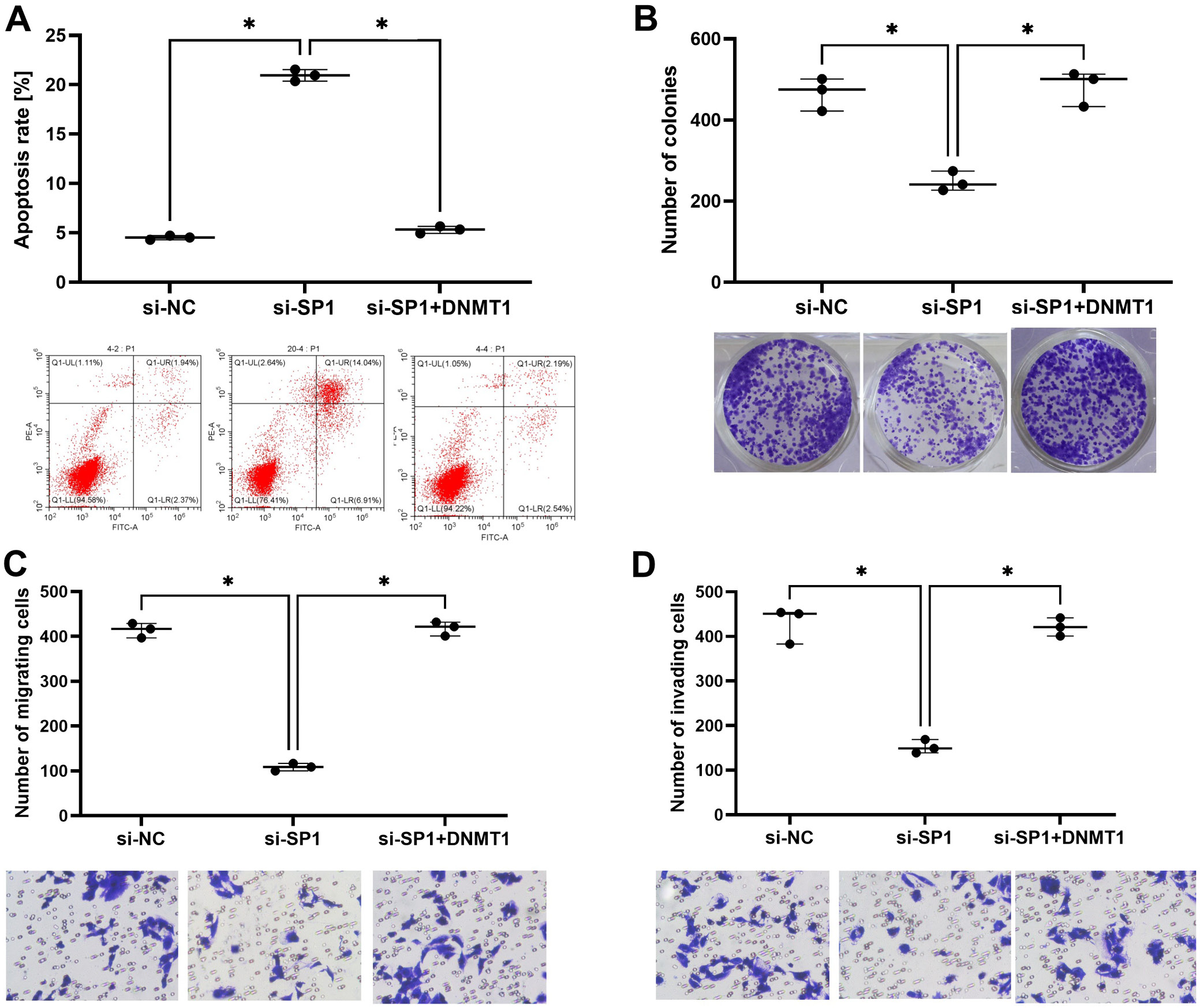
Downregulation of SP1 induced cell apoptosis in HTR-SV/neo cells but the overexpression of DNMT1 could counteract this (Figure 2A). Colony formation results showed that the number of colonies was decreased by SP1 knockdown but was restored by the overexpression of DNMT1 (Figure 2B). Cell migration and invasion were also inhibited by SP1 knockdown and restored by DNMT1 overexpression (Figure 2C,D).
miR-204-5p inhibited SP1/DNMT1 by targeting SP1 in HTR-8/SVneo cells
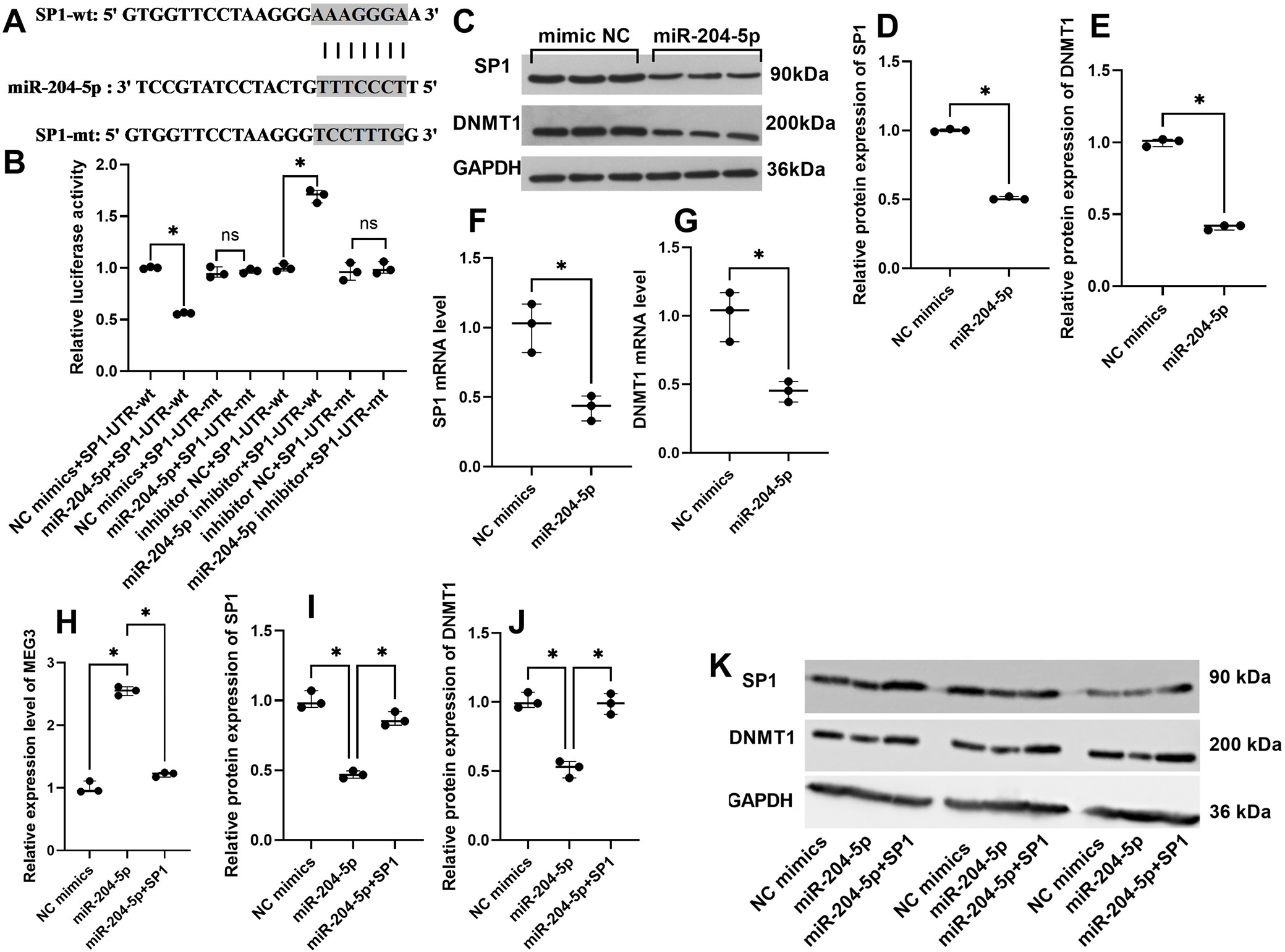
Based on predictions from the Starbase database, miR-204-5p is predicted to bind to SP1, and the putative binding sites are illustrated in Figure 3A. The cells were transfected with miR-204-5p mimics (miR-204-5p) or its negative control (NC mimics), miR-204-5p inhibitor or its negative control (inhibitor NC), and the pmirGLO plasmids containing the SP1-UTR-wt/mt. Dual luciferase reporter gene assays were performed to analyze the relative luciferase activity, and results showed that the relative luciferase activity was significantly reduced in cells co-transfected with miR-204-5p and SP1-UTR-wt and increased in cells transfected with miR-204-5p inhibitor and SP1-UTR-wt (Figure 3B). In addition, miR-204-5p upregulation inhibited SP1 and DNMT1 in mRNA and protein levels (Figure 3C–G). Conversely, overexpression of miR-204-5p upregulated MEG3 expression, an effect that was reversed by overexpression of SP1 (Figure 3H). The inhibitory effect of miR-204-5p on SP1/DNMT1 could be reversed by overexpression of SP1 (Figure 3I–K).
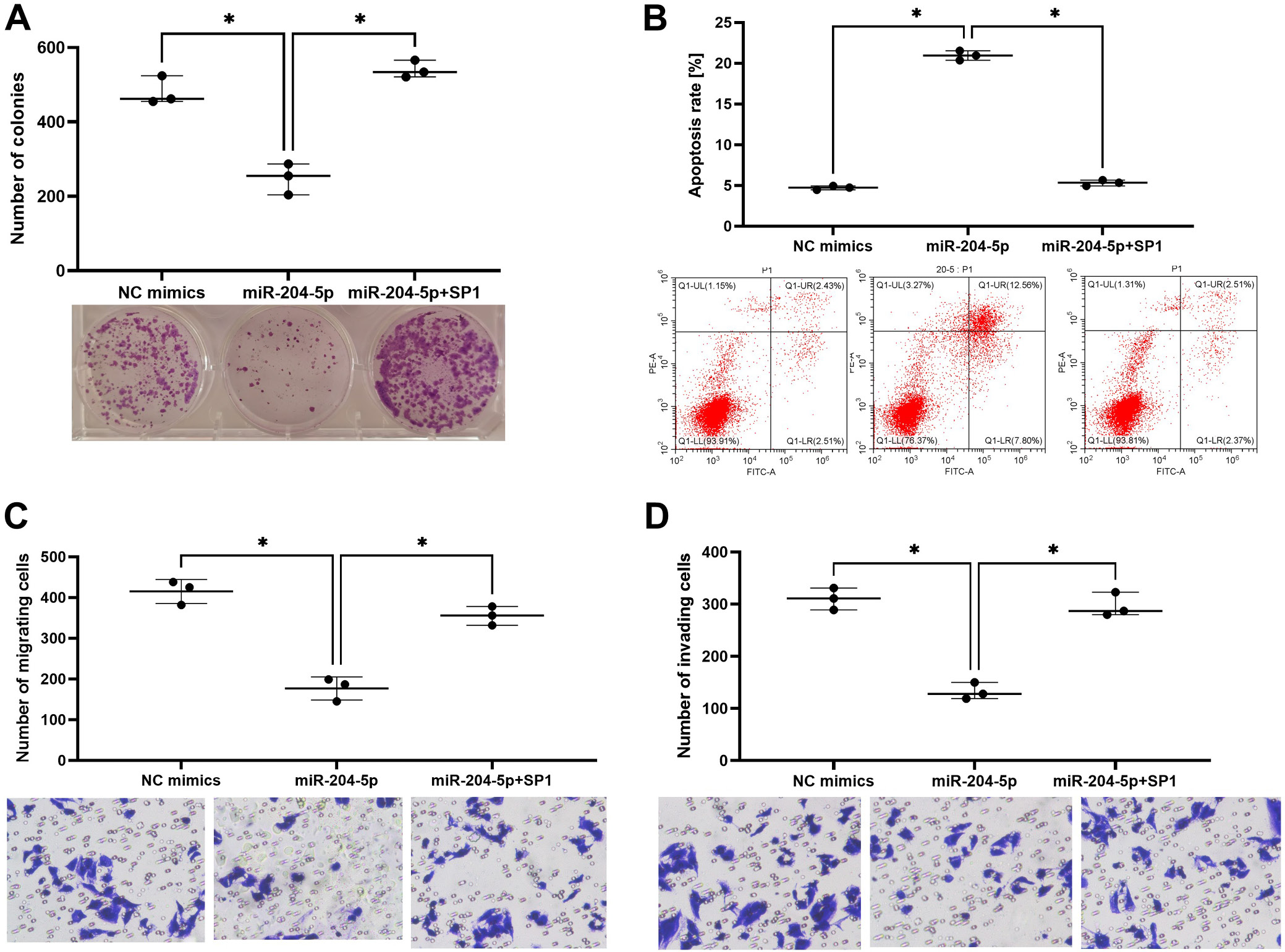
miR-204-5p regulated the invasion and migration of trophoblast cells by targeting SP1
Overexpression of miR-204-5p inhibited cell proliferation, invasion and migration but the inhibitory effect was recovered by SP1 overexpression in HTR-8/SVneo cells (Figure 4A,C,D). On the contrary, miR-204-5p overexpression enhanced apoptosis rates and SP1 overexpression reversed this (Figure 4B).
Discussion
This study advances the understanding of epigenetic regulation in RM. By elucidating the miR-204-5p/SP1/DNMT1/MEG3 axis, it provides insights into the molecular pathways contributing to trophoblast cell function and pregnancy maintenance. Specifically, it identifies miR-204-5p as a critical regulator of SP1 and DNMT1, influencing MEG3 expression and thereby affecting trophoblast cell behavior. These findings suggest potential therapeutic targets for managing RM, focusing on the modulation of miR-204-5p and its downstream effectors. In this study, we demonstrated that SP1 knockdown leads to a significant reduction in DNMT1 levels, which in turn upregulates MEG3 expression. This finding is consistent with existing literature that indicates SP1 enhances DNMT1 transcription.26 It was unveiled previously that protein expression levels of SP1 and DNMT1 were significantly low in human villous specimen from RM patients, in comparison with those from the patients with induced abortion or sporadic abortion; in vitro, knockdown of SP1 or DNMT1 could decrease the invasion and migration in trophoblast cells.27 In our study, the increase in apoptosis and decrease in migration and invasion following SP1 knockdown in trophoblast cells also reveal critical roles of SP1 in maintaining cell function. Further, these effects were reversed by DNMT1 overexpression, highlighting DNMT1’s role in cell proliferation and motility, and indicating that SP1 modulated the cell phenotypes via DNMT1. Such interactions have been well documented in cancer cells, where SP1 increases the activity of DNMT1, DNMT3A and DNMT3B, leading to hypermethylation of tumor suppressor gene.28, 29, 30 In our study, the decreased DNMT1 levels following SP1 knockdown suggest a similar regulatory mechanism in trophoblast cells, underscoring the broader implications of SP1-DNMT1 interactions beyond oncogenesis. In addition, the methylation inhibitor, 5-azadC, was used to induce the cells and we found that MEG3 expression was increased by 5-azadC, suggesting that MEG3 was regulated in trophoblast cells likely due to methylation. Furthermore, upregulation of SP1/DNMT1 in cells was found to partly reverse the change in MEG3 expression induced by 5-azadC. These results suggest that SP1/DNMT1 inhibited MEG3 expression via methylation regulation.
Previously, DNMT1 was identified to be essential for maintaining DNA methylation patterns during DNA replication, crucial for gene silencing and genomic stability.31, 32 For instance, in breast cancer, MEG3 expression was enhanced by knockdown of DNMT1 through demethylation of MEG3 promoter.23, 33 Our findings extend the interaction of DNMT1/MEG3 to trophoblast cells, suggesting that DNMT1-mediated methylation is vital for placental development and function.
A significant finding of this study is that miR-204-5p directly targets SP1, leading to decreased levels of both SP1 and DNMT1, which in turn upregulates MEG3 expression in trophoblast cells. This regulatory pathway was confirmed through dual-luciferase activity, RT-qPCR and western blot methods; miR-204-5p has been previously identified as a tumor suppressor in various cancers, regulating genes involved in proliferation and apoptosis.34, 35 Our study shows that miR-204-5p similarly impacts the apoptosis, invasion and migration of trophoblast cells, highlighting its role in reproductive health. MEG3 expression was increased by miR-204-5p overexpression, which was reversed by SP1 overexpression, indicates a complex regulatory mechanism involving miR-204-5p, SP1 and DNMT1 in trophoblast cells.
Limitations
Despite the significant findings, this study has limitations. First, the research was conducted in vitro using HTR-8/SVneo trophoblast cells, which may not fully replicate in vivo environment of placental tissue. Second, although the study identifies miR-204-5p as a regulator of SP1 and DNMT1, the broader regulatory network involving other miRNAs and transcription factors was not explored. Additionally, the clinical relevance of these findings needs further validation through studies involving human study participants and clinical samples.
Conclusions
The elucidation of the miR-204-5p/SP1/DNMT1/MEG3 pathway offers new perspectives on the molecular mechanisms in regulating trophoblast cells. Future research should explore therapeutic interventions targeting this pathway to improve pregnancy outcomes in preclinical animal studies. Additionally, investigating other miRNAs and lncRNAs involved in trophoblast function could further enhance the understanding of the complex regulatory networks in placental development.
Supplementary data
The supplementary materials are available at https://doi.org/10.5281/zenodo.14043134. The package includes the following files:
Supplementary Table 1. The results of homogeneity of variance test.
Supplementary Table 2. Statistical methods (Mann–Whitney U test) and test results used.
Supplementary Table 3. Kruskal–Wallis test with Dunn’s post hoc and Bonferroni correction and test results used.
Data availability
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
Consent for publication
Not applicable.
Use of AI and AI-assisted technologies
Not applicable.